Important Update:
The GRDI's Antimicrobial Resistance 2 (AMR2) project has been renamed to Antimicrobial Resistance – One Health (AMR-OH). This change reflects our commitment to a comprehensive approach to antimicrobial resistance, integrating human, animal and environmental health.
Funding period: 2022–2027
Leads: Tim McAllister, Agriculture and Agri-Food Canada, and Catherine Carrillo, Canadian Food Inspection Agency
Total GRDI funding: $9,800,000
Resistance to antimicrobial medicines, also known as antimicrobial resistance (AMR), is an important public health challenge. In particular, the development of resistance to antimicrobials by bacterial and fungal pathogens that were previously sensitive poses a great threat to human and animal health. Recognizing the complex nature of the AMR problem, the Genomics Research Development Initiative (GRDI) is funding a second interdepartmental project on antimicrobial resistance, AMR-OH.
The first GRDI AMR project focused on examining AMR in the context of agricultural production systems. Through work conducted in that project, researchers gained a better understanding of the critical processes contributing to the emergence of AMR in food production systems and identified key exposure pathways through which AMR bacteria of agricultural origin reach humans.
Building upon these results, the AMR-OH project will continue studying the risks associated with the emergence and dissemination of AMR. Federal genomics and regulatory communities interested in AMR have established the overarching goals for the project and identified an approach involving overlapping human, animal and environmental antimicrobial issues, known as the One Health approach, as the best approach for developing synergies among collaborating departments. Expanding the scope of this project will make it possible to address the interconnected nature of AMR transmission pathways, which reach across mandate-specific areas of responsibility. While the GRDI AMR-OH project will continue to study AMR transmission in the context of agri-food production systems, the scope of this project will also examine natural and wastewater systems, fisheries and health care environments.
Background
A total of 33 scientists and their teams from 6 federal science departments and agencies are contributing to the AMR-OH project. They are leveraging existing federal sampling programs and sample repositories to map the development and transit of antimicrobial resistance through health care, animals, plants, wastewater and the environment using genomics approaches, including metagenomics, whole genome sequencing and genomic epidemiology. The team is also developing world-class analytical tools and methods for genomics analysis. These are not only shared within the project, but also made widely available to academic and industry researchers in keeping with a commitment to open science. Similarly, the team regularly publishes and otherwise communicates their research findings, including making them available in academic databases to ensure rapid and wide distribution of the knowledge generated by the project.
The project seeks to develop and apply practical solutions for controlling and reducing AMR by investigating the relationships between AMR and antimicrobial use and by analyzing the movement of AMR across different nodes of the One Health continuum to better understand modes of transmission. The knowledge generated will be used to identify hotspots and intervention points and support evidence-based mitigation strategies, including informed health care and agri-food practices, regulations and policies as well as consumer education to protect vulnerable populations. The overriding goal is to preserve the effectiveness of the antimicrobials that Canadians rely on every day.
Figure 1: One Health AMR project
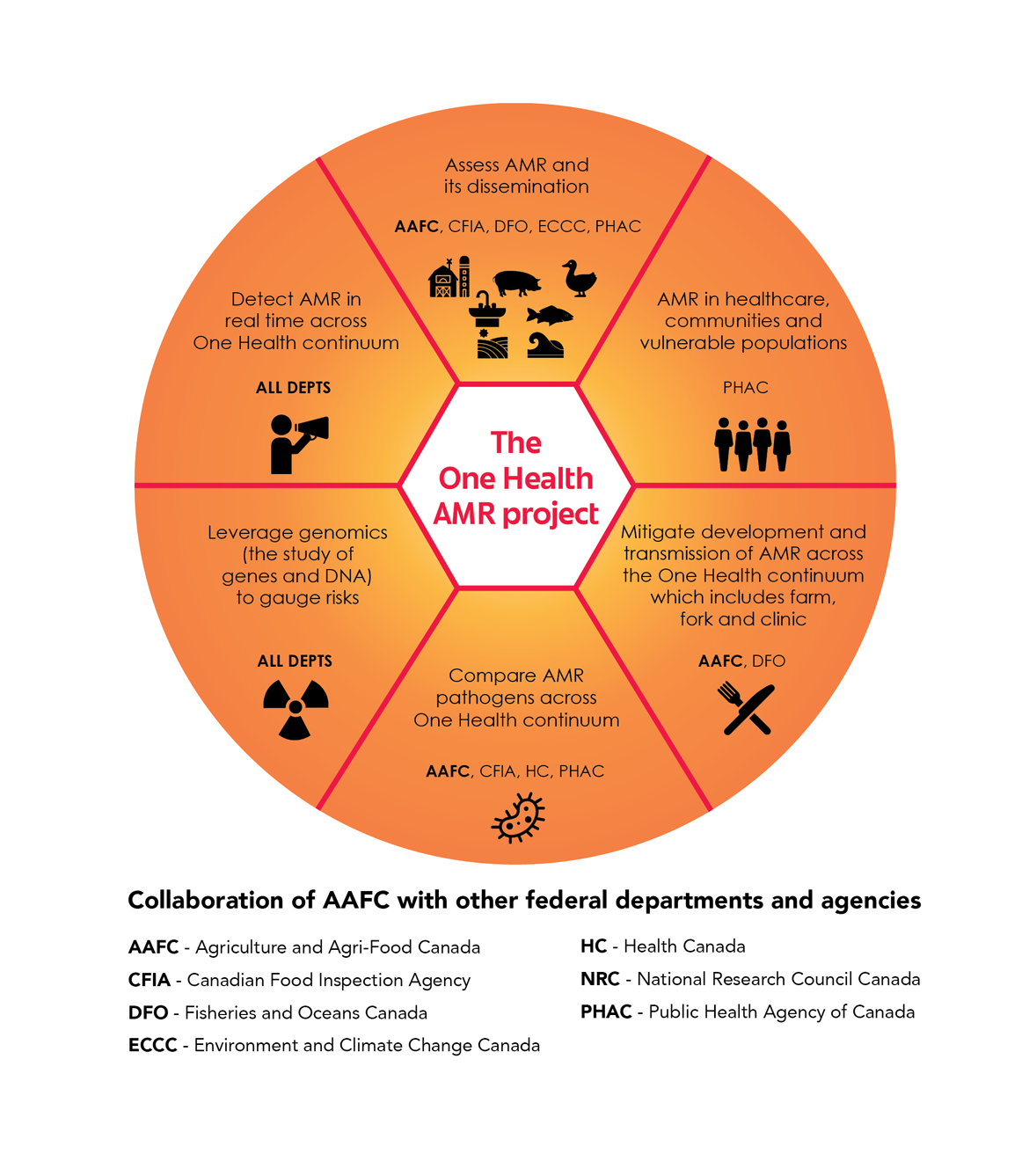
Figure 1: One Health AMR project - Text version
The One Health project on antimicrobial resistance (AMR) is organized into 6 themes. Each theme involves a mix of federal science-based departments and agencies collaborating to carry out work in different areas:
- Assess AMR and its dissemination in and among different environments, such as terrestrial and aquatic food production, agriculture and natural terrestrial and aquatic environments. Collaborators: Agriculture and Agri-Food Canada (lead), Canadian Food Inspection Agency, Fisheries and Oceans Canada, Environment and Climate Change Canada and Public Health Agency of Canada.
- Investigate AMR in healthcare settings, communities and vulnerable populations. Collaborator: Public Health Agency of Canada.
- Mitigate the development and transmission of AMR across the One Health continuum: farm to fork to clinic. Collaborators: Agriculture and Agri-Food Canada (lead) and Fisheries and Oceans Canada.
- Compare AMR pathogens across the One Health continuum. Collaborators: Agriculture and Agri-Food Canada (lead), Canadian Food Inspection Agency, Health Canada and Public Health Agency of Canada.
- Leverage genomics (the study of genes and DNA) to gauge risks. All collaborators: Agriculture and Agri-Food Canada, Canadian Food Inspection Agency, Fisheries and Oceans Canada, Environment and Climate Change Canada, Health Canada, NRC and Public Health Agency of Canada.
- Detect AMR in real time across the One Health continuum. All collaborators: Agriculture and Agri-Food Canada, Canadian Food Inspection Agency, Fisheries and Oceans Canada, Environment and Climate Change Canada, Health Canada, NRC and Public Health Agency of Canada.
Highlights
- 5-year collaboration
- 33 scientists and their teams
- 6 federal departments and agencies
Figure 2: AMR interactions and pathways

Figure 2: AMR interactions and pathways - Text version
A representation of bacterial cell, doubly depicted as an antibiotic capsule, showing the interactions between possible pathways for AMR transmission between aquatic and terrestrial environments. Overlayed on that background are 5 environmental areas as well as pathways mediating the exchange of AMR bacteria between them, all surrounded by representations of nature such as trees, deer, birds, lakes and mountains. There are many two-way solid and dotted arrows linking all environments in an elaborate, criss-crossing pattern, indicating complex interactions between them all. Each environment shows examples of its components in icon form with labels:
5 environments
- Aquatic: Aquaculture, fresh waters, ocean waters and wild fish
- Communities: Urban, suburban, vulnerable and rural
- Nosocomial: Hospital and clinic
- Waste: Manure, pesticides, sewage and residues like zinc and copper
- Farm: Livestock, produce, irrigation waters and imported foods
Highlighted pathways between environments
- Water supply
- Irrigation water
- Waste water
- Animal vectors: mammals, insects and birds
Objective
To identify intervention targets using a One Health approach by mapping the development and transit of antimicrobial resistance in health care settings, animals, plants, wastewater and the environment
Expected benefits
- Identification of intervention points, leading to improved surveillance practices and intervention strategies
- Reduction in spread of AMR to preserve the effectiveness of the antimicrobials that Canadians rely on every day
Areas of focus
Assessment of drivers, reservoirs and dissemination of AMR in environmental matrices
This area seeks to leverage the established scientific infrastructure, including rapidly advancing next generation sequencing and bioinformatics tools, as well as multi-agency expertise to investigate both the abundance and species distribution of AMR genes within different microbial communities (resistomes) across the One Health continuum.
Health care and Canadian communities, including vulnerable populations
This area seeks to identify AMR hotspots and intervention points in order to reduce AMR transmission between health care facilities, other at-risk populations and communities.
Mitigate development and transmission of AMR across the farm-to-fork-to-clinic continuum
This area aims to find solutions to reduce antimicrobial use in food production industries by optimizing existing practices and introducing new methods that reduce the need for antimicrobials. This applied research will strengthen management approaches that can have an impact on the bacterial community in food-animal production.
Phylogenomic comparison of AMR pathogens sourced from agricultural, environmental, food and clinical settings across Canada
This area aims to leverage expertise and resources from multiple departments to investigate AMR transmission between communities along the farm-to-fork-to-clinic continuum. The research outputs will inform the mapping of transmission pathways and identification of potential hotspots and high-risk bacterial species, thereby supporting the development of informed intervention strategies.
Leveraging genomics for risk science
This area seeks to integrate research data from the project in order to examine factors that contribute to increased risks of AMR to human, animal and environmental health via 2 avenues:
- Address specific knowledge gaps in processes that may be contributing to the dissemination of antibiotic resistant bacteria, antibiotic resistance genes and mobile genetic elements
- Develop risk science products (e.g. risk assessments, risk models) informed by genomic data generated through the project
Rapid detection and communication of AMR results across the One Health continuum
This area aims to ensure that all participants can readily access and accurately interpret all data, strains, reagents and insights.
Key deliverables
- Identification of AMR environmental hotspots and key drivers that will be prioritized for mitigation efforts
- Through molecular epidemiology, establishment of transfer pathways ("connecting the dots") between AMR in human clinical isolates and AMR bacteria in agricultural production systems, human communities and the environment
- Identification of vectors of AMR dissemination in natural and human-impacted terrestrial and aquatic environments
- Identification of the principal sources of AMR in human populations and the pathways of transmission between communities, health care facilities and other at-risk populations (e.g. long-term care settings, Northern and Indigenous communities, correctional institutions)
- Inclusion of genomic data in risk analyses and risk modelling to identify additional new approaches to mitigate AMR across the One Health continuum
- Identification of prospective studies to evaluate the impact of fungicides and antibiotics used in crop production on the development of AMR in environmental bacteria and fungi, which will provide regulatory authorities with data suitable for environmental risk assessments
- Efficacy of intervention strategies to reduce antimicrobial use in food production systems, to be validated under commercial conditions
- Assessment of the likelihood of exposure to AMR according to food production practices used by domestic and international producers, based on the surveillance of domestic and imported foods (raw and ready-to-eat meat as well as fresh and frozen produce)
- Genomics data for epidemiological and risk assessment tools to provide stronger evidence to support decision making in diverse settings, including terrestrial and aquatic food production systems and environmental management and health care settings
Funded participants
Collaborators
- BC Center for Aquatic Health Sciences
- Canadian Dairy Network
- Carleton University
- Dalhousie University
- Health Sciences Centre Winnipeg
- McGill University
- McMaster University
- Mount Sinai Hospital
- Simon Fraser University
- University of Alberta
- University of British Columbia
- University of Cambridge
- University of Guelph
- University of Lethbridge
- University of Prince Edward Island
- University of Saskatchewan
- University of Texas
- University of Waterloo
- University of Windsor
Publications
- Bradford, L., L. Yao, C. Anastasiadis, A. Cooper, B. Blais, A. Deckert, R. Reid-Smith, C. Lau, M. S. Diarra, C. Carrillo, and A. Wong, 2024. Limit of detection of Salmonella ser. Enteritidis using culture-based versus culture-independent diagnostic approaches. Microbiology Spectrum, https://doi.10.1128/spectrum.01027-24.
- Brown LP, Murray R, Scott A, Tien YC, Lau CHF, Tai V, Topp E. 2022. Responses of the soil bacterial community, resistome, and mobilome to a decade of annual exposure to macrolide antibiotics. Appl Environ Microbiol 6;88(8):e0031622. https://doi.org/10.1128/aem.00316-22
- Chongwu Y, Diarra MS, Rehman MA, Li L, Yu H, Yin X, Aslam M, CCarrillo CD, Yang C, Gong J. 2023. Virulence potential of antimicrobial resistant extraintestinal pathogenic E. coli from retail poultry meat in a Caenorhabditis elegans model. J. Food Protect. 86 (2023) 100008. https://doi.org/10.1016/j.jfp.2022.11.001
- Cooper AL, Low A, Wong A, Tamber S, Blais BW, Carrillo CD. 2024. Modeling the limits of detection for antimicrobial resistance genes in agri-food samples: a comparative analysis of bioinformatics tools. BMC Microbiol 24:31. https://doi.org/10.1186/s12866-023-03148-6
- Cooper AL, Wong A, Tamber S, Blais BW, Carrillo CD. 2024. Analysis of Antimicrobial Resistance in Bacterial Pathogens Recovered from Food and Human Sources: Insights from 639,087 Bacterial Whole-Genome Sequences in the NCBI Pathogen Detection Database. Microorganisms. 12(4):709. https://doi.org/10.3390/microorganisms12040709
- Das Q, Hasted TL, Lepp D, Yin X, Tang J, Chalmers G, Ross K, Boerlin P, Diarra MS. 2023. Transcriptional profiling of extraintestinal Escherichia coli exposed to cranberry pomace extracts alone or in combination with ceftriaxone. Front. Sustain. Food Syst. 6:957099. http://doi.org/10.3389/fsufs.2022.957099
- Demars M, McDowell T, Renaud J, Scott A, Fruci M, Topp E. 2024. Persistence and evidence for accelerated biodegradation of streptomycin in agricultural soils, Science of The Total Environment, 2024,172502,ISSN 0048-9697, https://doi.org/10.1016/j.scitotenv.2024.172502
- Diarra MS, Zhao X, Butaye PR. 2023. Antimicrobial use, antimicrobial resistance, and the microbiome in animals: volume II. Front. Vet. Sci. Volume 10. https://doi.org/10.3389/fvets.2023.1149010
- Diarra MS, Zhao X, Butaye PR, eds. 2023. Antimicrobial use, antimicrobial resistance, and the microbiome in animals, volume II. Lausanne: Frontiers Media SA. https://doi.org/10.3389/978-2-83251-839-7
- Flint A, Cooper A, Rao M, Weedmark K, Carrillo C, Tamber S. 2023. Targeted metagenomics using bait-capture to detect antibiotic resistance genes in retail meat and seafood. Front Microbiol. 2023 Jul 13;14:1188872. https://doi.org/10.3389/fmicb.2023.1188872
- Flint A, Laidlaw A, Li L, Raitt C, Rao M, Cooper A, Weedmark K, Carrillo C, Tamber S. 2022. Choice of DNA extraction method affects detection of bacterial taxa from retail chicken breast. BMC Microbiology. 22:230. https://doi.org/10.1186/s12866-022-02650-7
- Flores-Vargas G, Korber DR, Bergsveinson J. 2023. Sub-MIC antibiotics influence the microbiome, resistome and structure of riverine biofilm communities. Front. Microbiol. Sec. Aquatic Microbiology 31. https://doi.org/10.3389/fmicb.2023.1194952
- George PBL, Rossi F, St-Germain MW, Amato P, Badard T, Bergeron MG, Boissinot M, Charette SJ, Coleman BL, Corbeil J, Culley AI, Gaucher ML, Girard M, Godbout S, Kirychuk SP, Marette A, McGeer A, O'Shaughnessy PT, Parmley EJ, Simard S, Reid-Smith RJ, Topp E, Trudel L, Yao M, Brassard P, Delort AM, Larios AD, Létourneau V, Paquet VE, Pedneau MH, Pic É, Thompson B, Veillette M, Thaler M, Scapino I, Lebeuf M, Baghdadi M, Castillo Toro A, Cayouette AB, Dubois MJ, Durocher AF, Girard SB, Diaz AKC, Khalloufi A, Leclerc S, Lemieux J, Maldonado MP, Pilon G, Murphy CP, Notling CA, Ofori-Darko D, Provencher J, Richer-Fortin A, Turgeon N, Duchaine C. 2022. Antimicrobial Resistance in the Environment: Towards Elucidating the Roles of Bioaerosols in Transmission and Detection of Antibacterial Resistance Genes. Antibiotics (Basel). 11(7):974. https://doi.org/10.3390/antibiotics11070974
- Hetman BM, Pearl DL, Barker DOR, Robertson J, Nash JHE, Reid-Smith R, Agunos A, Carrillo C, Topp E, Van Domselaar G, Parmley EJ, Bharat A, Mulvey M, Allen V, Taboada EN. 2022. Combining analytical epidemiology and genomic surveillance to identify risk factors associated with the spread of antimicrobial resistance in Salmonella enterica subsp. enterica serovar Heidelberg. Microb Genom. 8(11):mgen000891. https://doi.org/10.1099/mgen.0.000891
- Isada MJ, Reist M, MacKinnon MC, Uhland FC, Young KM, Gibbens K, Parmley EJ, Carson CA. 2022. Characterisation of burden of illness measures associated with human (Fluoro)quinolone-resistant Campylobacter spp. infections - a scoping review. Epidemiol Infect. 150:e205. https://doi.org/10.1017/S095026882200139X
- Johnson L, Dufour S, Smith DDN, Manning A, Bulbul A, Binette S, Hamoutene D. 2023. Descriptive analyses of microbial communities in marine sediment microcosms spiked with fish wastes, emamectin benzoate, and oxytetracycline. Ecotoxicology and Environmental Safety, Volume 268, 115683, https://doi.org/10.1016/j.ecoenv.2023.115683
- Jonah L, Hamoutene D, Kingsbury M, Johnson L, Fenton AJ. 2024. A data compilation of antibiotic treatments in Canadian finfish aquaculture from 2016 to 2021 and the cumulative usage of antibiotics and antiparasitic drugs at marine sites. Environmental Reviews. https://doi.org/10.1139/er-2023-0124
- Kang H, Wang Q, Yu H, Guo Q, Weber L, Wu W, Lepp D, Cui SW, Diarra MS, Liu H, Shao S, Gong J. 2024. Validating the use of a newly developed cinnamaldehyde product in commercial broiler production. Poultry Science. https://doi.org/10.1016/j.psj.2024.103625
- Kang M, Charron P, Hoover E, Guan J, Firth I, Naushad I, and Huang H (correspondence). 2023. Complete genome sequence of a Canadian strain of Escherichia coli with multiple metal and antimicrobial resistance genes isolated from municipal biosolids. Microbiology Resources Announcement, ASM. 2023. Volume 12, Issue 5, e00083-23. https://doi.org/10.1128/mra.00083-23
- Kang M, Charron P, Hoover E, Huang H. 2024. Complete genome sequences of a Canadian strain of enteroaggregative Escherichia coli (EAEC) with multiple metals and antimicrobial resistance genes isolated from municipal waste-activated sludge. Microbiol Resour Announc 13:e01242-23. https://doi.org/10.1128/mra.01242-23
- Kang M, Naushad S, Hartke A, Firth I, Madey E, Ogunremi D, Huang H. 2022. Antibiotic resistomes and microbial communities in biosolid fertilizers collected from two Canadian wastewater treatment plants in a 10-years interval-potential risks to food chains? Front. Food. Sci. Technol. 2:894671. https://doi.org/10.3389/frfst.2022.894671
- Kujat Choy S, Neumann EM, Romero-Barrios P, Tamber S. 2024. Contribution of Food to the Human Health Burden of Antimicrobial Resistance. Foodborne Pathog Dis. 21(2):71-82. https://doi.org/10.1089/fpd.2023.0099
- Lau CHF, Capitani S, Tien YC. et al. 2024. Dynamic effects of black soldier fly larvae meal on the cecal bacterial microbiota and prevalence of selected antimicrobial resistant determinants in broiler chickens. Anim. microbiome 6:6. https://doi.org/10.1186/s42523-024-00293-9
- Lee C, Polo RO, Zaheer R, Van Domselaar G, Zovoilis A, McAllister T A. 2023. Evaluation of metagenomic assembly methods for the detection and characterization of antimicrobial resistance determinants and associated mobilizable elements. Journal of microbiological methods, 213, 106815. https://doi.org/10.1016/j.mimet.2023.106815
- Lee C, Zaheer R, Munns K, Holman DB, Van Domselaar G, Zovoilis A, McAllister TA. 2023. Effect of Antimicrobial Use in Conventional Versus Natural Cattle Feedlots on the Microbiome and Resistome. Microorganisms, 11(12), 2982. https://doi.org/10.3390/microorganisms11122982
- Lin, H. M. S. Diarra, G. Jia, X. Zhao. 2024. Detection in Salmonella from poultry: investigating the potential horizontal transfer of antimicrobial resistance and virulence genes. Poultry Science # PSJ-D-24-02029R1.
- Liu X, Floate KD,Gorzelak MA, Holman DB, Hrycauk S, Kubota H, Lupwayi N, Neilson JAD, Ortega PR, Petri RM, Tran L, Wang H, Wilches D, Yang X, Zorz J, Guarna MM. 2023. Prairie Agroecosystems: Interconnected Microbiomes of Livestock, Soil and Insects. Agriculture. 13:326. https://doi.org/10.3390/agriculture13020326
- Loest D, Uhland FC, Young KM, Li XZ, Mulvey MR, Reid-Smith R, Sherk LM, Carson CA. 2022. Carbapenem-resistant Escherichia coli from shrimp and salmon available for purchase by consumers in Canada: a risk profile using the Codex framework. Epidemiol Infect. 150:e148. https://doi.org/10.1017/S0950268822001030
- Lorenc, N., Leadbeater, S., Wang, J., Ronholm, J., Liu, X. 2024. A pilot study on the effects of in-feed probiotic Lactobacillus rhamnosus ATCC 53103 (LGG) on vaccinated Atlantic salmon (Salmo salar): Microbiomes and Aeromonas salmonicida challenge resilience. Can J Microbiol.
- Mak PHW, Rehman MA, Kiarie EG, Topp E, Diarra MS. 2022. Production systems and important antimicrobial resistant‑pathogenic bacteria in poultry: a review. Journal of Animal Science and Biotechnology. 13:148 https://doi.org/10.1186/s40104-022-00786-0
- McMahon TC, Kingombe CB, Mathews A, Seyer K, Wong A, Blais BW, Carrillo CD. 2022. Microbial Antagonism in Food-Enrichment Culture: Inhibition of Shiga Toxin-Producing Escherichia coli and Shigella Species. Frontiers in Microbiology 13: 880043. https://doi.org/10.3389/fmicb.2022.880043
- Muhammad B, Diarra MS, Islam MdR, Lepp D, Mastin-Wood RE, Topp E, Bittman S, Zhao X. 2022. Effects of litter from antimicrobial-fed broiler chickens on soil bacterial community structure and diversity. Can. J. Microbiol. 00: 1–11. https://doi.org/10.1139/cjm-2022-0086
- Nagarajan A, Goyette B, Raghavan V, Poulin-Laprade D, Rajagopal R. 2024. Integrating Anaerobic Digestion with Struvite Production for Enhanced Nutrient Recovery, Pathogen Reduction, and Circularity in Manure Management, Journal of Sustainable Agriculture and Environment, https://doi.org/10.1002/sae2.70018
- Oladeinde, A., K. Cook, A. Rehman, C. D. Carrillo, R. Woyda, C. Wiersma, Z. Abdo, J. Johnson Anna M. Bosch, M. Rothrock Jnr, and M. S. Diarra. 2024. Survival of Antimicrobial Resistant Salmonella Heidelberg Inoculated into Microcosms of Fresh Pine Wood Shavings for Broiler Litter. Can. J Microbiol. https://doi.10.1139/cjm-2024-0088.
- Phillips C, Chapman B, Agunos A, Carson CA, Parmley EJ, Reid-Smith RJ, Smith BA, Murphy CP. 2022. A scoping review of factors potentially linked with antimicrobial-resistant bacteria from turkeys (iAM.AMR Project). Epidemiol Infect. 150:e153. https://doi.org/10.1017/S0950268822001224
- Primeau CA, Bharat A, Janecko N, Carson CA, Mulvey M, Reid-Smith R, McEwen S, McWhirter JE, Parmley EJ. 2022. Integrated surveillance of extended-spectrum beta-lactamase (ESBL)-producing Salmonella and Escherichia coli from humans and animal species raised for human consumption in Canada from 2012 to 2017. Epidemiol Infect. 151:e14. https://doi.org/10.1017/S0950268822001509
- Robertson J, Schonfeld J, Bessonov K, Bastedo P, Nash JHE. 2023. A global survey of Salmonella plasmids and their associations with antimicrobial resistance. Microbial Genomics 9, 001002. https://doi.org/10.1099/mgen.0.001002
- Scott A, Murray R, Tien YC, Topp E. 2022. Contamination of hay and haylage with enteric bacteria and selected antibiotic resistance genes following fertilization with dairy manure or biosolids. Can J Microbiol. 68(4):249-257. https://doi.org/10.1139/cjm-2021-0326
- Shay JA, Haniford LSE, Cooper A, Carrillo CD, Blais BW, Lau, CH. 2023. Exploiting a targeted resistome sequencing approach in assessing antimicrobial resistance in retail foods. Environ. Microbiome 18(1):25. https://doi.org/10.1186/s40793-023-00482-0
- St-Laurent, R.E., Vincent, A. T., Paquet, V. E., Leduc, G. R., Lorenc, N.; Ronholm, J.; Liu, X.; Charette, S.J. 2024. Characterization of Aeromonas salmonicida mesophilic isolates from Alberta (Canada) allowing the development of a more sensitive Dictyostelium discoideum predation test. FEMS Microbiology. https://doi.org/10.1093/femsle/fnae078.
- Strong KM, Marasco KL, Invik J, Ganshorn H, Reid-Smith RJ, Waldner CL, Otto SJG, Kastelic JP, Checkley SL. 2023. Factors associated with antimicrobial resistant enterococci in Canadian beef cattle: a scoping review. 2023. Front Vet Sci. 10:1155772. https://doi.org/10.3389/fvets.2023.1155772
- Subirats J, Sharpe H, Tai V, Fruci M, Topp E. 2023. Metagenome meta-analysis reveals an increase in the abundance of some multidrug efflux pumps and mobile genetic elements in chemically polluted environments. Applied and Environmental Microbiology. 20:e01047-23. https://doi.org/10.1128/aem.01047-23
- Subirats J, Sharpe H, Topp E. 2022. Fate of Clostridia and other spore-forming Firmicute bacteria during feedstock anaerobic digestion and aerobic composting. Journal of Environmental Management 309: 114643. https://doi.org/10.1016/j.jenvman.2022.114643
- Subirats J, Sharpe H, Santoro D, Topp E. 2023. Modeling antibiotic concentrations in the vicinity of antibiotic-producing bacteria at the micron scale. Applied and Environmental Microbiology. 89(4):e0026123. https://doi.org/10.1128/aem.00261-23
- Tapp K, Deschênes M, Cooper A, Carrillo C, Blais B. 2023. Genomically informed custom selective enrichment of shiga toxigenic Escherichia coli (STEC) outbreak strains in foods using antibiotics. Journal of Food Protection 86(3):100052. https://doi.org/10.1016/j.jfp.2023.100052
- Tisza MJ, Smith DDN, Clark AE, Youn JH, NISC Comparative Sequencing Program, Khil PP, Dekker JP. 2023. Roving methyltransferases generate a mosaic epigenetic landscape and influence evolution in Bacteroides fragilis group. Nat Commun 14. https://doi.org/10.1038/s41467-023-39892-6
- VanderBurgt JT, Harper O, Garnham CP, Kohalmi SE, Menassa R. 2023. Plant production of a virus-like particle-based vaccine candidate against porcine reproductive and respiratory syndrome. Front. Plant Sci. 14:1044675. https://doi.org/10.3389/fpls.2023.1044675
- Venkatesan, M., Fruci, M, Verellen, LA, Skarina, T., Mesa, N., Flick, R., Stogios, P, and Savchenko A. 2023. Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics. Nature Communication. https://doi.org/10.1038/s41467-023-39778-7
- Waliaula, P. K., Kiarie, E. G. and Diarra M, S. 2024. Predisposition factors and control strategies of avian pathogenic Escherichia coli in laying hens. Front. Vet. Sci. 11:1474549. https://doi.10.3389/fvets.2024.1474549.
- Yang C, Das Q, Rehman MA, Yin X, Shay J, Gauthier M, Lau CHF, Ross K, Diarra MS. 2023. Microbiome of Ceca from Broiler Chicken Vaccinated or Not against Coccidiosis and Fed Berry Pomaces. Microorganisms. 11:1184. https://doi.org/10.3390/microorganisms11051184
- Yao L, Cooper AL, Gill A, et al. 2024. Overcoming Microbial Inhibition of S. Sonnei Through the Exploitation of Genomically Predicted Antibiotic Resistance Profiles for the Development of Food Enrichment Media. Journal of Food Protection. 2024 Jul;87(7):100302. DOI: 10.1016/j.jfp.2024.100302. PMID: 38754553
- Yilmaz G, Chan M, Lau CH, Capitani S, Kang M, Charron P, Hoover E, Topp E, Guan J. 2024. How Gut Microbiome Perturbation Caused by Antibiotic Pre-Treatments Affected the Conjugative Transfer of Antimicrobial Resistance Genes. Microorganisms. 12(11):2148. doi: 10.3390/microorganisms12112148.
- Young KM, Isada MJ, Reist M, Uhland FC, Sherk LM, Carson CA. 2022. A scoping review of the distribution and frequency of extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in shrimp and salmon. Epidemiol Infect. 151:e1 https://doi.org/10.1017/S0950268822001819
- Zaidi SE, Zaheer R, Barbieri R, Cook SR, Hannon SJ, Booker CW, Church D, Van Domselaar G, Zovoilis A, McAllister TA. 2022. Genomic Characterization of Enterococcus hirae From Beef Cattle Feedlots and Associated Environmental Continuum. Front Microbiol. 13:859990. https://doi.org/10.3389/fmicb.2022.859990
- Zaidi SE, Zaheer R, Poulin-Laprade D, Scott A, Rehman MA, Diarra M, Topp E, Domselaar GV, Zovoilis A, McAllister TA. 2023. Comparative Genomic Analysis of Enterococci across Sectors of the One Health Continuum. Microorganisms. 11(3):727. https://doi.org/10.3390/microorganisms11030727
- Zaidi SE, Zaheer R, Thomas K, Abeysekara S, Haight T, Saville L, Stuart-Edwards M, Zovoilis A, McAllister TA. 2023. Genomic Characterization of Carbapenem-Resistant Bacteria from Beef Cattle Feedlots. Antibiotics (Basel, Switzerland), 12(6), 960. https://doi.org/10.3390/antibiotics12060960
- Zhang L, Said LB, Hervé N, Zirah S, Diarra MS, Fliss I. 2022. Effects of drinking water supplementation with Lactobacillus reuteri, and a mixture of reuterin and microcin J25 on the growth performance, caecal microbiota and selected metabolites of broiler chickens. Journal of Animal Science and Biotechnology. 13:34. https://doi.org/10.1186/s40104-022-00683-6
- Zhang L, Said LB, Diarra MS, Fliss I. 2022. Effects of bacterial derived antimicrobial solutions on shelf-life, microbiota and sensory attributes of raw chicken legs under refrigerated storage condition, International Journal of Food Microbiology 389: 109958. https://doi.org/10.1016/j.ijfoodmicro.2022.109958
Contact us
Genomics R&D Initiative
Email: info@grdi-irdg.collaboration.gc.ca